About us
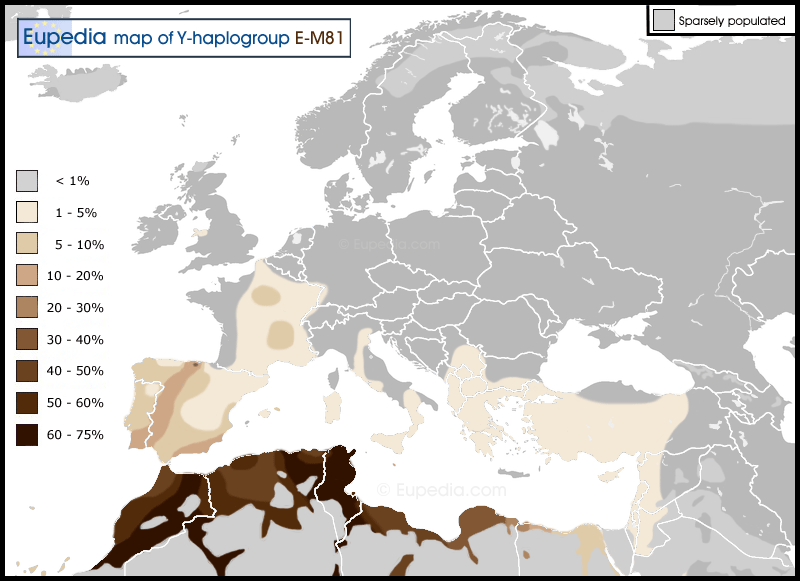
The Y haplogroup of the Rodríguez paternal line is E1b1b1b1a1 (Group D9): E-M35 (Elmenteitan) > E-M81/183 (Berber) > E-Z5009 (but Z5013/MZ131/MZ141-) (Masmuda), a moroccan tribe around Marrakesh. This tribe, with the leader Tarif, leaded a group of Zenata tribe in the Umayyad conquest of Hispania. After the conquest, Tarif lost his power over the Zenatas and decided to found the Masmuda kingdom of Barghawata, because he came from Barbate (Spain). After, the Sinhala tribe, named themselves as Almoravids, conquered the Masmuda capital Aghmat and founded a new one for their new kingdom, Marrakesh. But Masmudas, named themselves as Almohads, founded a new capital in the Atlas mountains, Tinmed, and finally they conquered the Almoravids capital, Marrakesh, and founded the Almohad Caliphate with a new capital, Seville (Spain). Finally, when moriscos were expelled from Spain, he probably served to Rodrigo and adopted the surname Rodriguez to avoid the expulsion. This agrees also with A2 paternal blood group (see below).
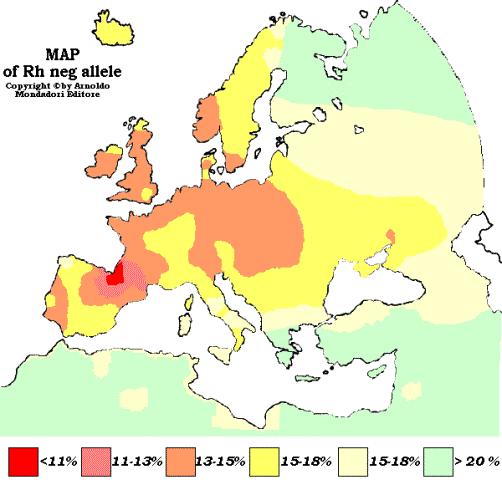
The Y haplogroup of the Peña paternal line is R1b1a1a2a1a2a4: R1b-M269 (Anatolian) > R1b-P312 (Beaker Folk: Rhenish) > R1b-DF27/ZZ19 (Gascon) > R1b-A431/A432 (Basque), where it took place the current mutation. The greatest pool is in the Pyrenees, which is concidental with the frankish Hispanic March, and this agrees with Oto de la Peña, navarre knight of frankish origin who took his surname from the village Peña in Navarre and was the founder of San Juan de la Peña monastery near Jaca (Aragon). Moreover, the eight-pointed star in his coat of arms is coincidental with the frankish Navarre symbol and the coat of arms of Estella town.

The mtDNA haplogroup of the Peña (Castañeda) maternal line is U5a1, which was originated in Pitted Ware Culture in Götaland and this haplogroup was found after in Baden-Württemberg and Luxembourg. Together with autosomal analysis, this agrees with visigoth occupation of "Gothic Plains", currently the "Land of Fields" between Valladolid and Palencia provinces, where the additional HVR1 mutation 16169T appeared. It also maches with a matrilineal CCR5 del32 mutation of nordic origin.
You can check this U5a1 haplogroup by using the "Compare" button in mitosearch.org with the user BXPRW and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16169T 16192T 16256T 16270T 16399G 73G 263G 309.1C 315.1C
The mtDNA haplogroup of the Valdivieso (Casas) maternal line of my father is U5b1c1a, which was originated from Carni people in Friuli (Italy) and it was found after in Aquitani people (Gascon in France and Basques in Spain). Together with homozygous RHD gene deletion of my father (i4001527), this agrees with a basque ancestry of maternal line of my father, and also points out to basque ancestry of some paternal ancestors of my father, as Zarraga lineage from Oñate (Gipuzkoa) and Sarria lineage (toponymic surname from Biscay).
You can check this U5b1c1a haplogroup by using the "Compare" button in mitosearch.org with the user W5DEV and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16183C 16189C 16270T 16311C 16336A 53A 55A 73G 150T 263G 309.1C 315.1C
The haplogroups U5 (Ursula clan) was originated in Yamnaya culture (Ukraine) in the easter European steppe and in western Europe they splited in two branches (a and b) in the migrations that brought Indo-European languages to western Europe.

The mtDNA haplogroup of the Peña (Castro Gutiérrez) maternal line is H20a (Helena clan a), which was originated in the Near Est. This agrees with Sephardic migration in Roman empire through Sicily, where the additional HVR1 mutation 16298C appeared.
You can check this H20a haplogroup by using the "Compare" button in mitosearch.org with the user 2Y99Z and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16218T 16298C 16328A 16362C 263G 309.1C 309.2C 315.1C (Image from PLoS ONE. 2008; 3(4): e2062)

Autosomal ancestry:
Eye Colors (predicted by GEDmatch):
Me: high melanin with weak amber gradient: rs1129038=CC (high melanin), rs4778241=AA (medium melanin), rs1470608=TT (large amounts of melanin), rs2240203=CC (large amounts of melanin), rs1105879=AA (weak amber gradient), rs3947367=TT (no contrasted sphincter), rs4779685=TC (no flecks/nevi), rs11634406=GG (no flecks/nevi).
Father: medium melanin with weak amber gradient and contrasted sphincter: rs1129038=CC (high melanin), rs4778241=AA (medium melanin), rs1470608=GT (medium melanin), rs2240203=CT (medium melanin), rs1105879=AA (weak amber gradient), rs3947367=CT (contrasted sphincter), rs4779685=CC (no flecks/nevi), rs11634406=GG (no flecks/nevi)
Mother: high melanin with weak amber gradient: rs1129038=CC (high melanin), rs4778241=AC (high melanin), rs1470608=TT (large amounts of melanin), rs2240203=CC (large amounts of melanin), rs1105879=AA (weak amber gradient), rs3947367=TT (no contrasted sphincter), rs4779685=TC (no flecks/nevi), rs11634406=GG (no flecks/nevi)
Father's mother: medium melanin with contrasted sphincter and flecks/nevi: rs1129038=CT (medium melanin, yellowish), rs4778241=AC (high melanin), rs1470608=GT (medium melanin), rs2240203=CT (medium melanin), rs1105879=AC (no amber gradient), rs3947367=CT (contrasted sphincter), rs4779685=CC (flecks/nevi), rs11634406=AG (flecks/nevi)
Common blood groups: Colton(a) (rs28362692 = CC), Yt(a) (rs1799805 = GG), Lutheran(b) (rs28399653 = GG), Auberger(a) (rs1135062 = AA), Memphis(-) (rs5036 = TT) and Diego(b) (rs2285644 = GG).
Knops: Kn(a) (rs41274768 = GG), McC(a) (rs17047660 = AA) and SI(a) (rs17047661 = AA)
Kell: k (rs8176058 = GG), Kp(b) (rs8176059 = GG) and Js(b) (rs8176038 = AA)
Particular blood groups:
Duffy: all Fy(ab) (rs12075 = GA) except me Fy(b) (rs12075 = AA). Fy(a) with rs12075 = G is Germanic

Kidd: all Jk(ab) (rs1058396 = GA) except me Jk(b) (rs1058396 = AA). Jk(a) with rs1058396 = G came from Africa: Berber?
Lewis: all Le(b) (rs601338 = CC : Secretory FUT2 / rs812936 = AA + rs778986 = GG : Active FUT3) except father's mother Le(ab) (rs601338 = TC). Le(a) with rs601338 = T came from Africa: Berber?
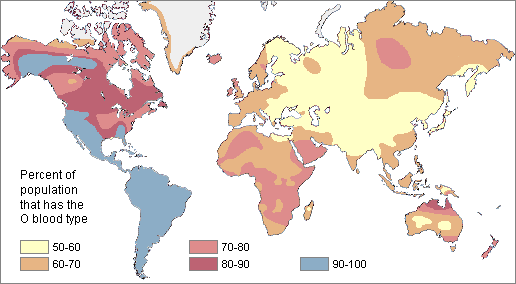
ABO: all have "no-B" SNPs: rs7853989 GG + rs8176743 + rs8176749 + rs8176722 + rs41302905 CC
Me: A2O1 (rs1053878 + rs8176704 AG (A2) / rs687289 AG (+/O) + rs512770 GG (O1))
Father: A1/A2 (rs1053878 + rs8176704 GA (A1/A2) / rs687289 = AA (no O) + rs512770 GG (O1))
Mother: O1/O2 (rs1053878 + rs8176704 GG (A1) / rs687289 = GG (O) + rs512770 GA (O1/O2))
Father's mother: O1/O2 (rs1053878 + rs8176704 GG (A1) / rs687289 = GG (O) + rs512770 GA (O1/O2))
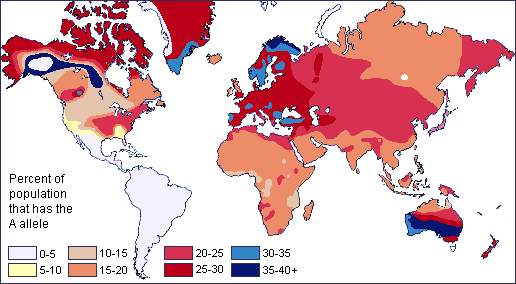
A2 with rs1053878 = A and O2 with rs512770 A came from North Africa (probably Berber/Masmuda paternal ancestry and Berber/Sinhaja maternal ancestry)
Another isoforms:
CYP3A4: rs4986910 = AA, except my mother and me rs4986910 = AG (1A/3). The SNP rs4986910 = G (CYP3A4*3) agrees with germanic ancestry of maternal line.
APOE: rs7412 = CC (epsilon3), except my father and me rs7412 = TC (epsilon2/epsilon3). The Alzheimer's protective SNP rs7412 = T (epsilon2) agrees with hungarian ancestry of Villarragut family, who came from avars kings (Hungarian plate) and took refuge in Vilagut (Catalonia, ancient carolingian Hispanic March) (article in Spanish): http://www.revclinesp.es/es/longevidad-genotipo-apoe-2-3-una/articulo/S0014256510003085/
PON1: mother rs662 = TC (QR) and father and me rs662 = CC (RR). The atherosclerosis' protective SNP rs662 = C (Q192R) agrees with berber ancestry of both maternal and paternal line: https://deepblue.lib.umich.edu/bitstream/handle/2027.42/46312/210_2003_Article_833.pdf;jsessionid=0BE972A344440463CD7C9D9093FBD45A?sequence=1
CETP: The hypercholesterolemia's protective SNP rs5882 = G (I405V) and SNPs rs5883 T + rs9930761 C (delta9) agrees with berber/sinhaja ancestry of my maternal line:
Me: Wt/I405V (rs5882 = AG (Wt/I405V) / rs5883 CC + rs9930761 TT (no delta9))
Father: Wt (rs5882 = AA (Wt) / rs5883 CC + rs9930761 TT (no delta9))
Mother: I405V/I405V-delta9 (rs5882 = GG (I405V) / rs5883 CT + rs9930761 TC (Wt/delta9))
Father's mother: Wt/I405V-delta9 (rs5882 = AG (Wt/I405V) / rs5883 CT + rs9930761 TC (Wt/delta9))
We have almost a complete Homo sapiens sapiens ancestry (obtained with Ancient calculator tool):
- Me: 99,66% H. s. sapiens, 0,25% H. s. neanderthalensis and 0,09% H. s. denisoviensis
- Father: 99,63% H. s. sapiens, 0,21% H. s. neanderthalensis and 0,16% H. s. denisoviensis
- Mother: 99,74% H. s. sapiens, 0,13% H. s. neanderthalensis and 0,13% H. s. denisoviensis
- Father's mother: 99,4% H. s. sapiens, 0,19% H. s. neanderthalensis and 0,41% H. s. denisoviensis
The following ancestral composition was obtained with autosomal files by using Ancient Ancestry tool:
Me:

Father:

Mother:

Mother's father:


The Y haplogroup of the Peña paternal line is R1b1a1a2a1a2a4: R1b-M269 (Anatolian) > R1b-P312 (Beaker Folk: Rhenish) > R1b-DF27/ZZ19 (Gascon) > R1b-A431/A432 (Basque), where it took place the current mutation. The greatest pool is in the Pyrenees, which is concidental with the frankish Hispanic March, and this agrees with Oto de la Peña, navarre knight of frankish origin who took his surname from the village Peña in Navarre and was the founder of San Juan de la Peña monastery near Jaca (Aragon). Moreover, the eight-pointed star in his coat of arms is coincidental with the frankish Navarre symbol and the coat of arms of Estella town.

The mtDNA haplogroup of the Peña (Castañeda) maternal line is U5a1, which was originated in Pitted Ware Culture in Götaland and this haplogroup was found after in Baden-Württemberg and Luxembourg. Together with autosomal analysis, this agrees with visigoth occupation of "Gothic Plains", currently the "Land of Fields" between Valladolid and Palencia provinces, where the additional HVR1 mutation 16169T appeared. It also maches with a matrilineal CCR5 del32 mutation of nordic origin.
You can check this U5a1 haplogroup by using the "Compare" button in mitosearch.org with the user BXPRW and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16169T 16192T 16256T 16270T 16399G 73G 263G 309.1C 315.1C
The mtDNA haplogroup of the Valdivieso (Casas) maternal line of my father is U5b1c1a, which was originated from Carni people in Friuli (Italy) and it was found after in Aquitani people (Gascon in France and Basques in Spain). Together with homozygous RHD gene deletion of my father (i4001527), this agrees with a basque ancestry of maternal line of my father, and also points out to basque ancestry of some paternal ancestors of my father, as Zarraga lineage from Oñate (Gipuzkoa) and Sarria lineage (toponymic surname from Biscay).
You can check this U5b1c1a haplogroup by using the "Compare" button in mitosearch.org with the user W5DEV and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16183C 16189C 16270T 16311C 16336A 53A 55A 73G 150T 263G 309.1C 315.1C
The haplogroups U5 (Ursula clan) was originated in Yamnaya culture (Ukraine) in the easter European steppe and in western Europe they splited in two branches (a and b) in the migrations that brought Indo-European languages to western Europe.


The mtDNA haplogroup of the Peña (Castro Gutiérrez) maternal line is H20a (Helena clan a), which was originated in the Near Est. This agrees with Sephardic migration in Roman empire through Sicily, where the additional HVR1 mutation 16298C appeared.
You can check this H20a haplogroup by using the "Compare" button in mitosearch.org with the user 2Y99Z and/or by using the on-line tool in dna.jameslick.com by pasting the following mutation in a TXT file and then by uploading this file: 16218T 16298C 16328A 16362C 263G 309.1C 309.2C 315.1C (Image from PLoS ONE. 2008; 3(4): e2062)
Autosomal ancestry:
Eye Colors (predicted by GEDmatch):
Me: high melanin with weak amber gradient: rs1129038=CC (high melanin), rs4778241=AA (medium melanin), rs1470608=TT (large amounts of melanin), rs2240203=CC (large amounts of melanin), rs1105879=AA (weak amber gradient), rs3947367=TT (no contrasted sphincter), rs4779685=TC (no flecks/nevi), rs11634406=GG (no flecks/nevi).
Father: medium melanin with weak amber gradient and contrasted sphincter: rs1129038=CC (high melanin), rs4778241=AA (medium melanin), rs1470608=GT (medium melanin), rs2240203=CT (medium melanin), rs1105879=AA (weak amber gradient), rs3947367=CT (contrasted sphincter), rs4779685=CC (no flecks/nevi), rs11634406=GG (no flecks/nevi)
Mother: high melanin with weak amber gradient: rs1129038=CC (high melanin), rs4778241=AC (high melanin), rs1470608=TT (large amounts of melanin), rs2240203=CC (large amounts of melanin), rs1105879=AA (weak amber gradient), rs3947367=TT (no contrasted sphincter), rs4779685=TC (no flecks/nevi), rs11634406=GG (no flecks/nevi)
Father's mother: medium melanin with contrasted sphincter and flecks/nevi: rs1129038=CT (medium melanin, yellowish), rs4778241=AC (high melanin), rs1470608=GT (medium melanin), rs2240203=CT (medium melanin), rs1105879=AC (no amber gradient), rs3947367=CT (contrasted sphincter), rs4779685=CC (flecks/nevi), rs11634406=AG (flecks/nevi)
Common blood groups: Colton(a) (rs28362692 = CC), Yt(a) (rs1799805 = GG), Lutheran(b) (rs28399653 = GG), Auberger(a) (rs1135062 = AA), Memphis(-) (rs5036 = TT) and Diego(b) (rs2285644 = GG).
Knops: Kn(a) (rs41274768 = GG), McC(a) (rs17047660 = AA) and SI(a) (rs17047661 = AA)
Kell: k (rs8176058 = GG), Kp(b) (rs8176059 = GG) and Js(b) (rs8176038 = AA)
Particular blood groups:
Duffy: all Fy(ab) (rs12075 = GA) except me Fy(b) (rs12075 = AA). Fy(a) with rs12075 = G is Germanic
Kidd: all Jk(ab) (rs1058396 = GA) except me Jk(b) (rs1058396 = AA). Jk(a) with rs1058396 = G came from Africa: Berber?
Lewis: all Le(b) (rs601338 = CC : Secretory FUT2 / rs812936 = AA + rs778986 = GG : Active FUT3) except father's mother Le(ab) (rs601338 = TC). Le(a) with rs601338 = T came from Africa: Berber?
ABO: all have "no-B" SNPs: rs7853989 GG + rs8176743 + rs8176749 + rs8176722 + rs41302905 CC
Me: A2O1 (rs1053878 + rs8176704 AG (A2) / rs687289 AG (+/O) + rs512770 GG (O1))
Father: A1/A2 (rs1053878 + rs8176704 GA (A1/A2) / rs687289 = AA (no O) + rs512770 GG (O1))
Mother: O1/O2 (rs1053878 + rs8176704 GG (A1) / rs687289 = GG (O) + rs512770 GA (O1/O2))
Father's mother: O1/O2 (rs1053878 + rs8176704 GG (A1) / rs687289 = GG (O) + rs512770 GA (O1/O2))
A2 with rs1053878 = A and O2 with rs512770 A came from North Africa (probably Berber/Masmuda paternal ancestry and Berber/Sinhaja maternal ancestry)


Another isoforms:
CYP3A4: rs4986910 = AA, except my mother and me rs4986910 = AG (1A/3). The SNP rs4986910 = G (CYP3A4*3) agrees with germanic ancestry of maternal line.
APOE: rs7412 = CC (epsilon3), except my father and me rs7412 = TC (epsilon2/epsilon3). The Alzheimer's protective SNP rs7412 = T (epsilon2) agrees with hungarian ancestry of Villarragut family, who came from avars kings (Hungarian plate) and took refuge in Vilagut (Catalonia, ancient carolingian Hispanic March) (article in Spanish): http://www.revclinesp.es/es/longevidad-genotipo-apoe-2-3-una/articulo/S0014256510003085/
PON1: mother rs662 = TC (QR) and father and me rs662 = CC (RR). The atherosclerosis' protective SNP rs662 = C (Q192R) agrees with berber ancestry of both maternal and paternal line: https://deepblue.lib.umich.edu/bitstream/handle/2027.42/46312/210_2003_Article_833.pdf;jsessionid=0BE972A344440463CD7C9D9093FBD45A?sequence=1
CETP: The hypercholesterolemia's protective SNP rs5882 = G (I405V) and SNPs rs5883 T + rs9930761 C (delta9) agrees with berber/sinhaja ancestry of my maternal line:
Me: Wt/I405V (rs5882 = AG (Wt/I405V) / rs5883 CC + rs9930761 TT (no delta9))
Father: Wt (rs5882 = AA (Wt) / rs5883 CC + rs9930761 TT (no delta9))
Mother: I405V/I405V-delta9 (rs5882 = GG (I405V) / rs5883 CT + rs9930761 TC (Wt/delta9))
Father's mother: Wt/I405V-delta9 (rs5882 = AG (Wt/I405V) / rs5883 CT + rs9930761 TC (Wt/delta9))
We have almost a complete Homo sapiens sapiens ancestry (obtained with Ancient calculator tool):
- Me: 99,66% H. s. sapiens, 0,25% H. s. neanderthalensis and 0,09% H. s. denisoviensis
- Father: 99,63% H. s. sapiens, 0,21% H. s. neanderthalensis and 0,16% H. s. denisoviensis
- Mother: 99,74% H. s. sapiens, 0,13% H. s. neanderthalensis and 0,13% H. s. denisoviensis
- Father's mother: 99,4% H. s. sapiens, 0,19% H. s. neanderthalensis and 0,41% H. s. denisoviensis
The following ancestral composition was obtained with autosomal files by using Ancient Ancestry tool:
Me:

Father:

Mother:

Mother's father:
