About us
A ‘‘Copernican’’ Reassessment of the Human Mitochondrial DNA Tree from its Root
Doron M. Behar, Mannis van Oven, et. al., The American Journal of Human Genetics 90, 675–684, April 6, 2012 675,
Opinion D.K. from ISOGG forum (Message #27816), (I am agree with this opinion- SJP):
This is a landmark paper. mtDNA results will now be classified in a completely new way. Currently we are given our "differences" from the Revised Cambridge Reference Sequence which was the first mitochondrial genome to be sequenced. This sequence belongs to haplogroup H and is a very recent branch of the mtDNA tree.
The authors have now effectively reconstructed the mtDNA haplotype ... the common maternal ancestor of all humans, which they've called
the Reconstructed Sapiens Reference Sequence (RSRS).
Administrator of this Baltic Sea Project is intended to involve this new step of our knowledge to information about DNA Full Mitochondrial Sequence of the project participants'. Administrator will indicate when had occurred splitting the lines of evolution main groups of Project participants.
Stanislaw J Plewako
Administrator of this
Baltic Sea DNA Project
Current phylogenetic trees of humankind is available according Mannis van Oven here:
http://www.phylotree.org/Y/tree/index.htm (paternal line, based on Y-DNA mutations analysis)
http://www.phylotree.org/tree/main.htm (maternal line, based on mtDNA mutations analysis)
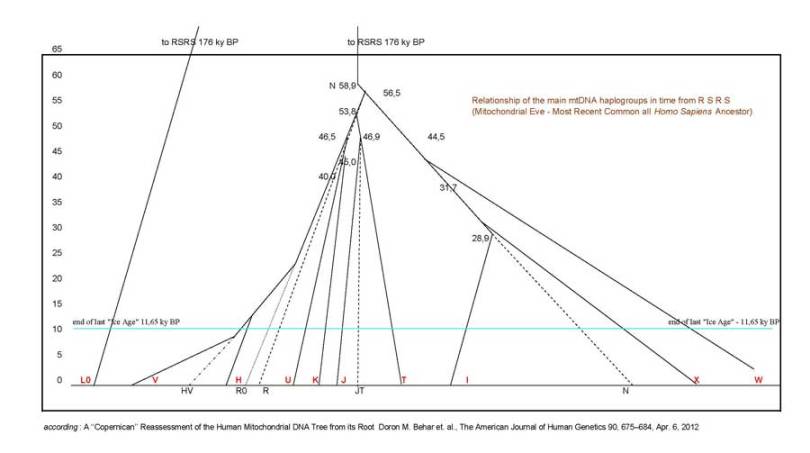
Estimated time (years before present) of mtDNA phylogenetic tree splits for branches presented in Baltic Sea DNA Project
split times and structure according: A ‘‘Copernican’’ Reassessment of the Human Mitochondrial DNA Tree from its Root
all dating from appendix to this work, see: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3322232/
0. RSRS (=Reconstructed Sapiens Reference Sequence, Mitochondrial Eve) have lived in Africa ~200.000 ybp
see: https://isogg.org/wiki/Most_recent_common_ancestor#Matrilineal_MRCA
1. L3 arise in Africa ~65.250 BC (or between 62.828 ybp and 71.696 ybp)
(RSRS > L1-6 > L3)
10. L3* branch
11. N branch 58.859 ybp (L3 -> N)
1102. S split up ybp
111. N1a split up ~18.117 ybp
111. N1b1a split up ~8.987 ybp
1111. N1a1b2 =I haplogroup ~20.860 ybp
1111. I1 split up ~15.231 ybp
1111. I1a1a split up ~3.327 ybp
1111. I1a1a3a terminal subclade
1111. I2 split up ~6.387 ybp
1112. N1b1a split up ~6.980 BC (L3 -> N)
1121. W5 split up ~11.075 ybp
1121. W6a split ~6.445 ybp
113. X2 split up ~19.234 ybp
113. X2e1b PERFECT MATCH terminal subclade split up ~500-1000 ybp
115041. V 9.739 ybp (N -> R -> HV -> V)
115041. V1a1a split up ~2.681 ybp
115041. V2a1a split up ~968 ybp
115041. V3 split up ~6.838 ybp
115042. H 12.846 ybp (N -> R -> HV -> H)
115042. H1 split up ~9.889 ybp
115042. H1a split up ~6.307 ybp
115042. H1a2 split up ~2.909 ybp
115042. H1a3a split up ~3.859 ybp
115042. H1b split up ~6.237 ybp
115042. H1b1 split up ~4.955 ybp
115042. H1b2 split up ~3.662 ybp
115042. H1c split up ~6.448 ybp
115042. H1c3 split up ~4.532 ybp
115042. H1c3b split up ~1.031 ybp terminal subclade
115042. H1e split up ~8.502 ybp
115042. H1e2 split up ~3.508 ybp
115042. H1h1 split up ~4.318 ybp
115042. H1i1 split up ~2.734 ybp
115042. H1n split up ~8.886 ybp
115042. H1n4 split up ~4.174 ybp
115042. H1u split up ~6.990 ybp
115042. H2 split up ~ 11.905 ybp
115042. H2a1 split up ~7.713 ybp
115042. H2a1a split up ~6.044 ybp
115042. H2a1c split up ~1.785 ybp
115042. H3 split up ~8.919 ybp
115042. H3h1 split up ~1.569 ybp
115042. H3r split up ~3.646 ybp
115042. H4 split up ~10.617 ybp
115042. H4a1a1a split up ~5.891 ybp
115042. H5 split up ~9.877 ybp
115042. H5a split up ~9.111 ybp
115042. H5a1a split up ~2.977 ybp
115042. H6 split up ~10.946 ybp
115042. H6a split up ~9.504 ybp
115042. H6a1a split up ~7.140 ybp
115042. H6a1a2a split up ~3.980 ybp
115042. H6a1a4 split up ~4.135 ybp
115042. H7 split up ~8.891 ybp
115042. H7a split up ~7.382 ybp
115042. H7b split up ~4.066 ybp
115042. H8 split up ~8.341 ybp
115042. H8c split up ~6.400 ybp
115042. H10 split up ~8.595 ybp
115042. H10e split up ~4.689 ybp
115042. H11 split up ~8.425 ybp
115042. H11a split up ~6.122 ybp
115042. H11a2a2 split up ~1.304 ybp terminal subclade
115042. H13 split up ~12.476 ybp
115042. H13b1 split up ~8.074 ybp
115042. H15 split up ~8.693 ybp
115042. H15a1 split up ~6.136 ybp
115042. H54 split up ~8.271 ybp
115042. H80 split up ~1.735 ybp terminal subclade
115042. H85 split up ~3.738 ybp
1151. U2e1 split up ~14.898 ybp
1151. U3 split up ~32.703 ybp
1151. U3b1b split up ~1.892 ybp terminal subclade
1151. U4 split up ~17.493 ybp
1151. U4a split up ~14.950 ybp
1151. U4b1a2 split up ~1.781 ybp
1151. U5 split up ~30.248 ybp
1151. U5a1 split up ~16.936 ybp
1151. U5a1a1 split up ~6.836 ybp
1151. U5a1b1h terminal subclade
1151. U5a1c1 split up ~4.990 ybp
1151. U5a2 split up ~18.416 ybp
1151. U5a2a1 split up ~6.094 ybp
1151. U5b1b1a split up ~2.281 ybp
1151. U5b1b1a1a split up ~1.356 ybp
1151. U5b1b2 split up ~1.981 ybp
1151. U5b1e1a split up ~985 ybp PERFECT MATCH terminal subclade
1151. U5b2 split up ~20.040 ybp
1151. U7 split up ~18.052 ybp
1151. U8 split up ~43.034 ybp
1151. U8a1a1b1 terminal subclade
11511. U8b2 = K haplogroup 26.681 ybp (N -> R -> U -> K)
11511. K1a split up ~18.433 ybp
11511. K1a4a1 split up ~9.617 ybp
11511. K1a4a1a2b split up ~1.808 ybp PERFECT MATCH terminal subclade
11511. K1c split up ~13.935 ybp
11511. K1c1c split up ~1.508 ybp
11511. K2a split up ~9.105 ybp
11511. K2b1 split up ~6.961 ybp
119. N9 split up 43.700 BC (N → N9)
119. N9c = Y haplogroup split up 24.576 ybp
153. JT 46.908 ybp (N -> R -> JT)
1531. J 34.258 ybp (N -> R -> JT -> J)
1531. J1b1a1 split up ~6.971 ybp
1531. J1c split up ~13.072 ybp
1531. J1c10a PERFECT MATCH terminal subclade
1531. J1c2 split up ~9.762 ybp
1531. J1c3 split up ~8.855 ybp
1531. J1c5 split up ~10.632 ybp
1531. J2b1 split up ~9.282 ybp
1531. J2b1a split up ~7.518 ybp
1532. T 25.149 ybp (N -> R -> JT -> T)
1532. T1 split up ~16310 ybp
1532. T1a1 split up ~6.997 ybp
1532. T1a1b split up ~3.864 ybp
1532. T2 split up ~19.316 ybp
1532. T2a1a split up ~6.426 ybp
1532. T2b split up ~10.070 ybp
1532. T2b4a split up ~3.573 ybp
1532. T2f split up ~11.260 ybp
12. M branch 49.590 ybp (L3 -> M)
120. M* including Q
121. CZ split up ~36.473 ybp (L3 -> M -> CZ)
1211. C split up ~23.912 ybp (CZ -> C)
1211. C1b split up ~14.794 ybp
1212. Z split up ~21.662 ybp (CZ -> Z)
1212. Z1a1a split up ~1.049 ybp
122. D split up ~38.434 ybp (L3-> M -> D)
122. D4 split up ~24.580 ybp
2. L1 arise in Africa 128.520 ybp (RSRS > L1'2'3'4'5'6 -> L1)
L1b split up ~4.334 ybp
Stanislaw J Plewako
Administrator of this Baltic Sea DNA Project
2017-07-02, modif. 2019-10-10